JBIMS AI in Microbiome Science Event Series
Upcoming Event:
AI × Microbiome Innovation | 3rd Annual JBIMS Microbiome Innovation Forum
- Date: Friday, May 8, 1 - 6pm
- Location: The David Brower Center
We are thrilled to announce the capstone event in the JBIMS AI in Microbiome Science Event Series: the third annual JBIMS Microbiome Innovation Forum (aka Industry Mixer) will take place on Friday, May 8 from 1-6PM at The Brower Center. This vibrant gathering is designed to connect local microbiome-related researchers, startup founders, investors, policy-makers, non-profit groups, and more to spark new collaborations and amplify the impact of our shared work. This year’s event will provide a forum for microbiome leaders to explore future directions of the field, in particular those that have been unleashed by the power of AI.
New this year, the mixer will be geared toward identifying bottlenecks in the translation of microbiome technologies, emphasizing those that AI can help resolve. Leveraging the diverse expertise this event attracts, we will moderate interactive discussions to identify possible solutions and potential partnerships. The mixer will utilize AI-facilitated networking, discussion, and synthesis to harness the creativity of individual contributors and diverse groups. We will
JBIMS AI in Microbiome Science Event Series
Upcoming Event: Workshop #2 - Advanced Applications and the Future of AI in Microbiome Science
- Date: Thursday, November 13, 1 - 5pm
- Location: Bakar BioEnginuity Hub
This workshop picks up where our first session left off - moving from the basics into more applied uses of AI for microbiome science. It’s open to everyone: whether you’re new to computational tools and want some guided practice, or you already have experience and are looking for advanced applications. We’ll work through hands-on examples, hear a set of short lightning talks on real research applications, and wrap up with a conversation on ethics, responsibility, and how to make sure these methods are used thoughtfully across different research contexts.
JBIMS AI in Microbiome Science Event Series
We are excited to announce two upcoming workshops to kick off the JBIMS AI in Microbiome Science event series. The workshops are a complementary pair designed to cultivate skills, understanding, and conversation around the use of AI in the field of microbiome science
Workshop #1: Foundations of AI for Microbiome Science
- Date: Wednesday, October 22, 1 - 5pm
- Location: The Faculty Club at UC Berkeley
- Description: This workshop will provide a practical introduction to essential AI concepts, data standardization, and application to microbiome research, targeting those new to the field.
Workshop #2: Advanced Applications and the Future of AI in Microbiome Science
- Date: Thursday, November 13, 1 - 5pm
- Location: Bakar BioEnginuity Hub
- Description: This workshop picks up where our first session left off - moving from the basics into more applied uses of AI for microbiome science. It’s open to everyone: whether you’re new to computational tools and want some guided practice, or you already have experience and are looking for advanced applications. We’ll work through hands-on examples, hear a set of short lightning talks on real research applications,
UPCOMING EVENT: JBIMS Industry Mixer 2025

We are excited to announce our second annual JBIMS Industry Mixer, this year sponsored by Switch Bioworks! The mixer will be a vibrant gathering designed to connect local academic researchers and microbiome companies, spark new collaborations, and amplify the impact of our shared work.
Location: Bakar BioEngenuity Hub at UC Berkeley
Date: May 15th, 2025
Time: 1:00 - 6:00 PM
This year’s mixer will feature a dynamic lineup of activities aimed at fostering meaningful connections between academia and industry, including:
- Keynote talks from distinguished leaders across the microbiome science landscape, including Dr. Jay Keasling.
- Poster presentations and lightning talks highlighting cutting-edge research
- A panel discussion featuring local organizations driving innovation in microbiome science and biotechnology.
- Talks on entrepreneurship, offering insights into launching successful start-ups from academic roots.
- Recruitment opportunities for Bay Area scientists at all career stages
Last year’s event drew over 120 attendees, representing 23 companies and many research groups from UC Berkeley, LBNL, UCSF, UC Davis, SFSU, and more.
This is a unique opportunity to network with peers, forge new partnerships, and engage with the next
COBRA improves viral MAGs
Drs. LinXing Chen and Jill Banfield (IGI and EPS departments at UC Berkeley) unveiled Contig Overlap Based Re-Assembly (COBRA), a bioinformatic tool that resolves viral MAG assembly breakpoints. They tested the tool using >200 published freshwater metagenomes and found that COBRA vastly improved our ability to generate circular genome assemblies from 34% to 70%.
Published February 6, 2024 in Natural Microbiology.

Created by LinXing Chen.
The Swiss Army Knife of post-fire Paraburkholderia: rhamnolipid methyl esters
Mira Liu and members of the noteworthy Traxler Lab (PMB Department, UC Berkeley) found that multiple Paraburkholderia spp. isolated from burned soils produce unusual rhamnolipid surfactants. Upon further experimentation, they found that these molecules may serve multiple functions: they can inhibit other post-fire bacteria and fungi, aid in bacterial motility, and potentially solubilize hydrocarbons found in char. Published February 5, 2024 in ISME.
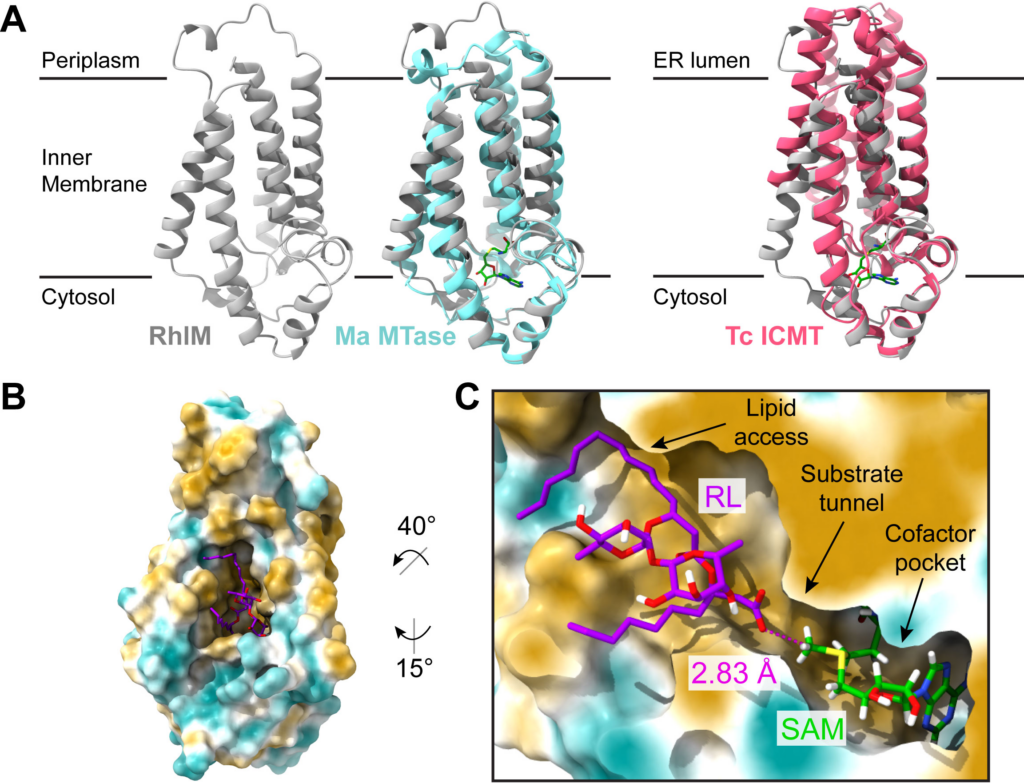
From Liu et al. (2024)
Predicting rhizosphere microbiome dynamics
Dr. Gianna Marschmann and members of LBNL's Ecology Department used genome-inferred models of microbial traits and energy budgets to successfully predict metabolic strategies of plant-associated microbes. In doing so, their models highlighted important life history traits and trade-offs, which provide useful insights for improving microbial components of global biogeochemical models. Published February 5, 2024 in Nature Microbiology
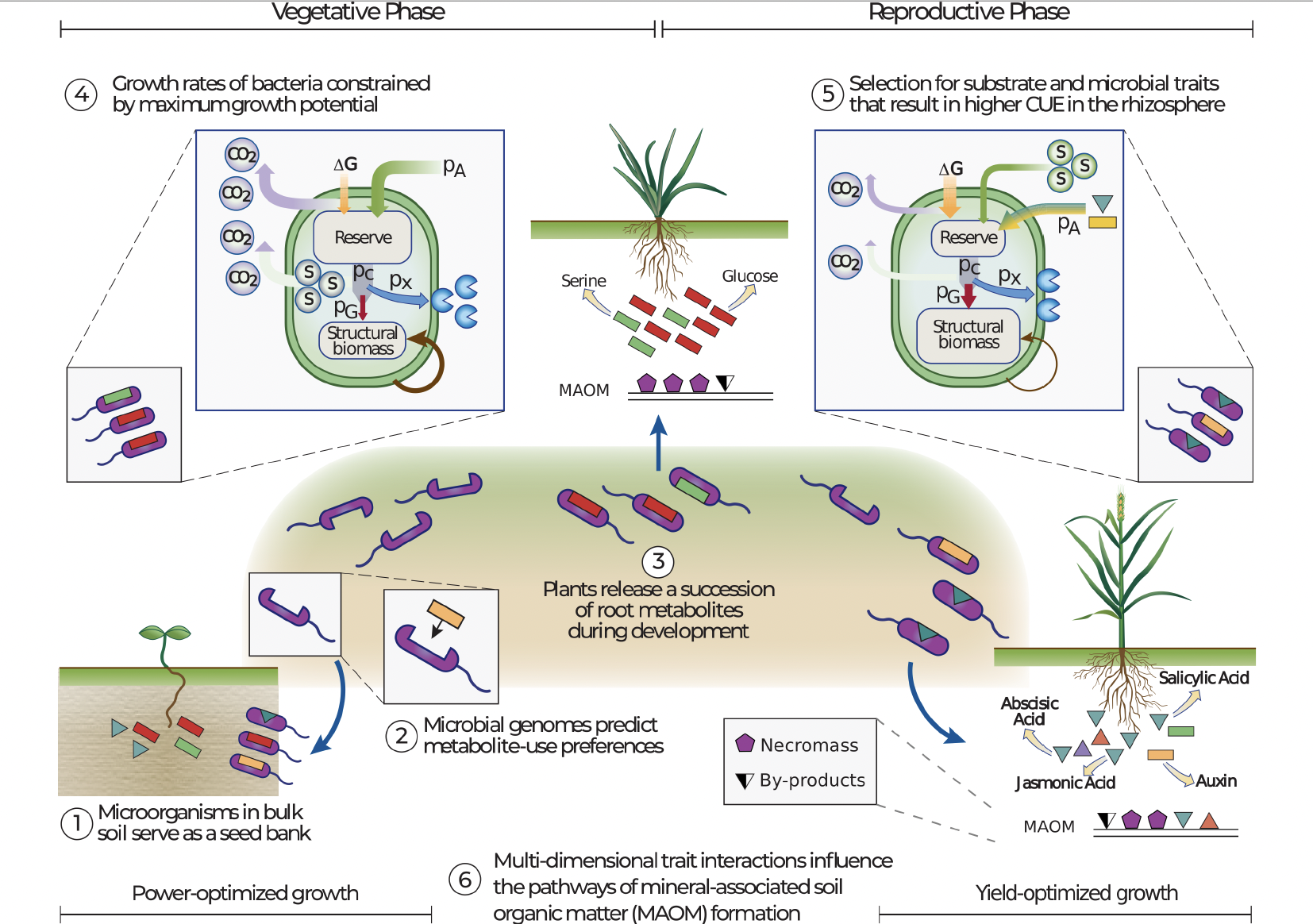
From Marschmann et al. (2024)
Harnessing fermentation in sulfate-reducing bioreactors
Fermentative microbes are often undesirable in sulfate-reducing bioreactors used for the bioremediation of acid mine drainage, due to their competition for organic substrates with sulfate reducers. Drs. Tomas Hessler and Jill Banfield along with collaborators from the University of Cape Town find bioreactor and metagenomic evidence that H2 generated by fermenters may greatly benefit sulfate reducers in the reactors and propose this syntrophy could be harnessed in the future. Published February 1, 2024 in Environmental Science and Technology.
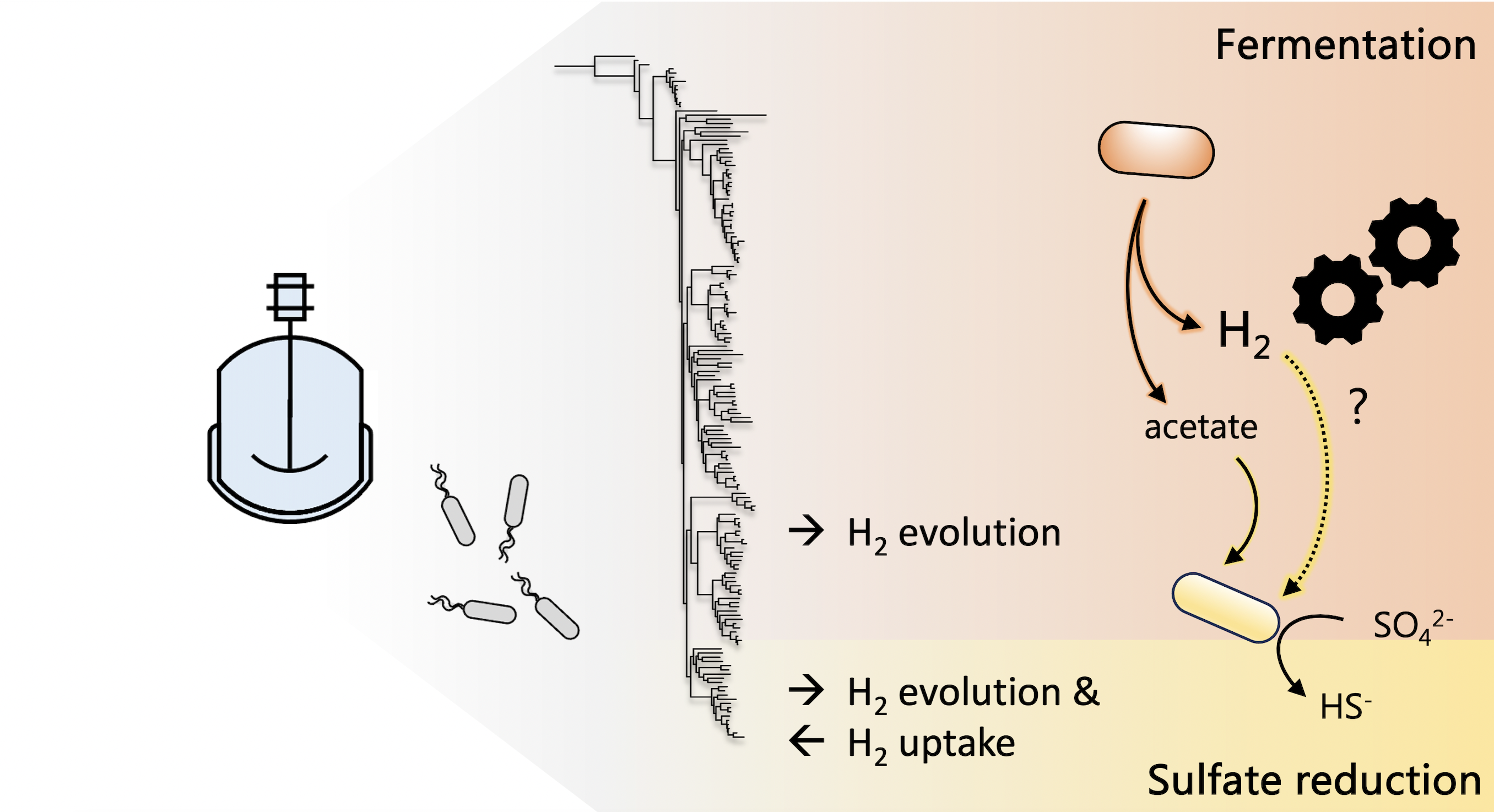
From Hessler et al. (2024)
Municipal water age affects resident microbiota
Dr. Hannah Greenwald Healy and members of the Nelson Research Group (Department of Civil and Environmental Engineering at UC Berkeley) found that microbial abundance increased more than an order of magnitude in a model water distribution system during 6 months in which water use was reduced due to COVID closures. The microbial community composition also changed over that period of time, and the shift depended on the community of initial constituents. The findings illustrate how tap water age influences resident microbial communities. Published January 22, 2024 in npj Biofilms and Microbiomes.
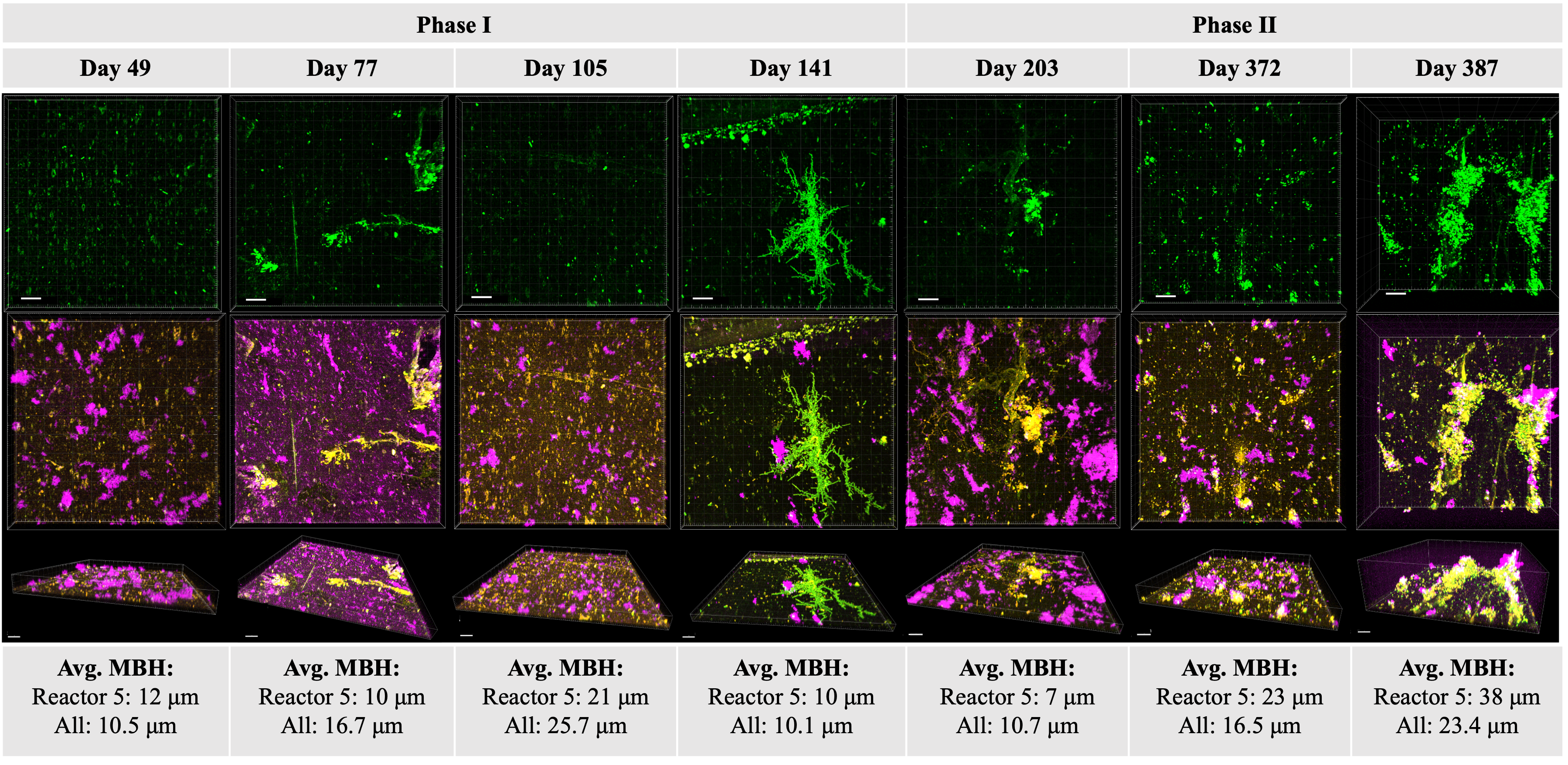
From Greenwald Healy et al. (2024)
Nitrogen status modulates root exudates
Using their highly-controlled EcoFab 2.0 plant growth device, Dr. Vlastimil Novak and researchers from the Northen Metabolomics Lab discovered that root exudates from Brachypodium distachyon depend on the plant’s nitrogen status. Such exudate changes, in turn, alter the growth of associated rhizobacteria. Published January 3, 2024 in Science Advances.

Photo credit: Thor Swift / Berkeley Lab
A guide to randomly barcoded transposon mutant libraries
A review by by Surya Tripathi and members of the Deutschbauer Lab offers a comprehensive guide to constructing randomly barcoded transposon mutant libraries for better understanding gut microbiota.
Published December 22, 2023 in Cell Reports.
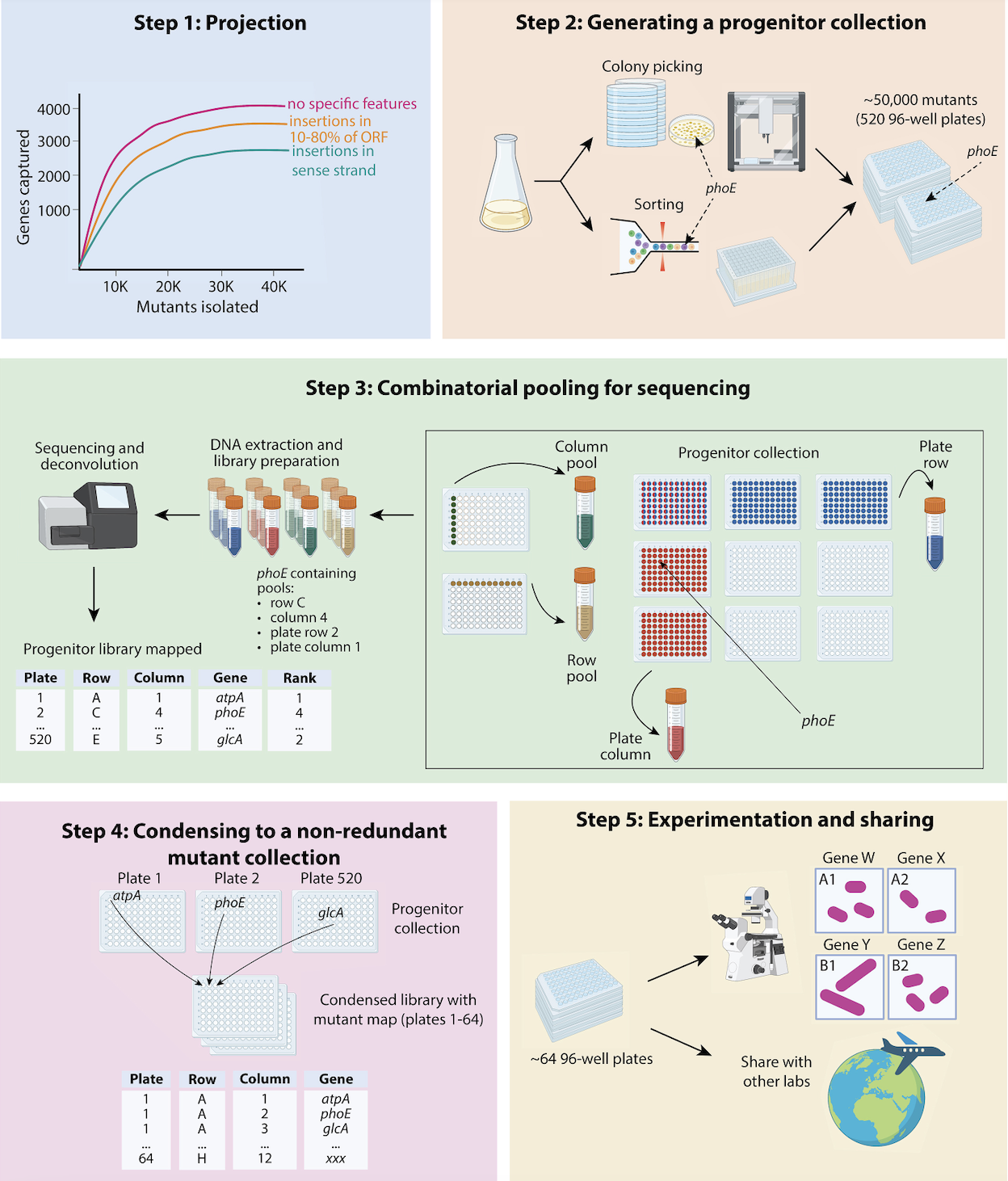
From Tripathi et al. (2024)
Establishment of the gut phageome
Dr. Yue Clare Lou and members of the Banfield Lab discovered patterns of bacteriophage establishment and succession in the human gut microbiome using fecal metagenomes from mothers and infants. Published December 13, 2023 in Cell Host & Microbe.
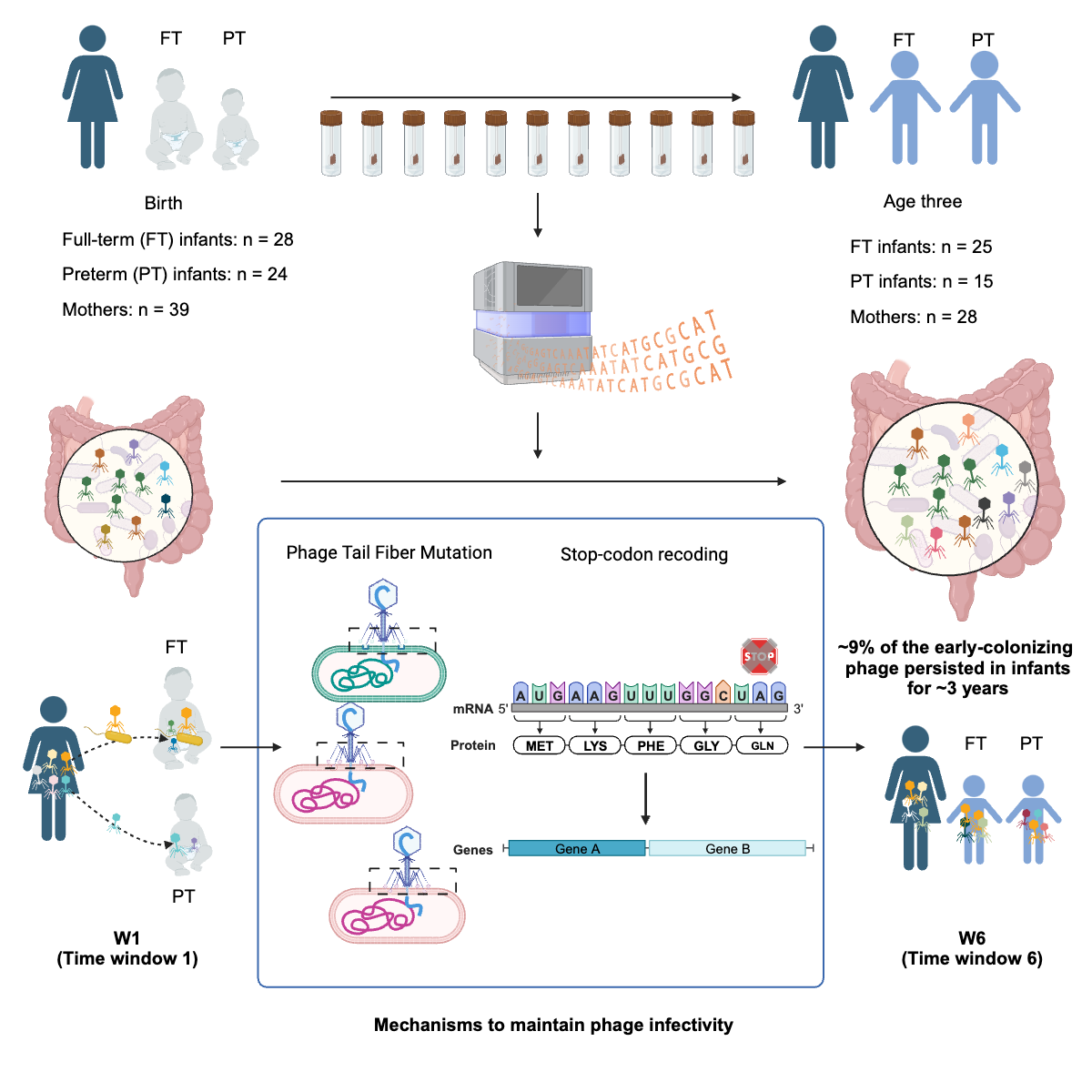
From Lou et al. (2024)
Bacterial sensory domains predict physical environment
Helen Park and members of the Environmental Genomics and Systems Biology at LBNL used machine learning to predict physical characteristics of the environment using bacterial sensory domains from a large database of metagenomic sequences. The work highlights the importance of sensory domains in the interaction between a bacterium and its environment. Published December 11, 2023 in mSystems.
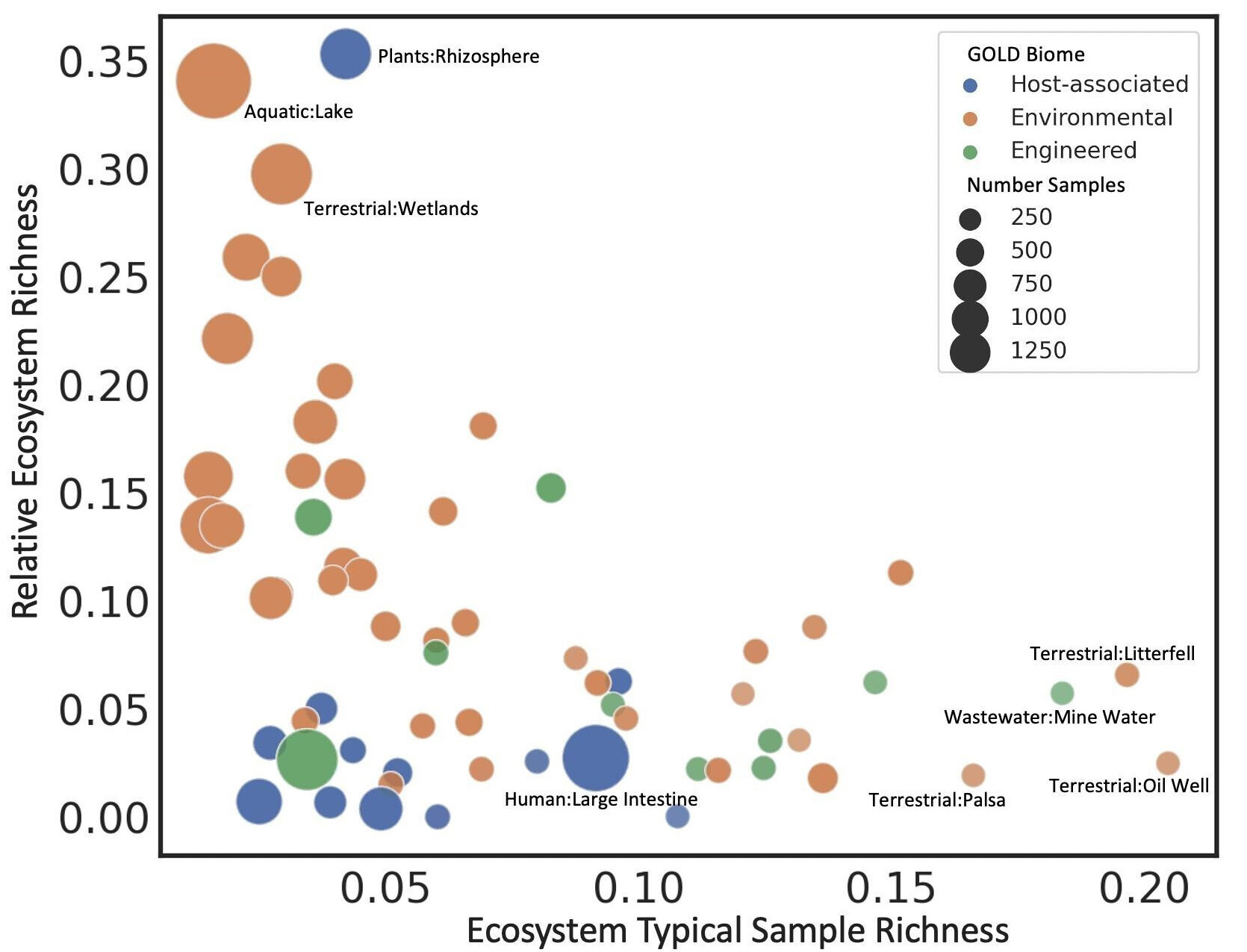
From Park et al. (2024)
Mapping gene essentiality in phages
Dr. Denish Piya and members of the Innovative Genomics Institute used a genome wide CRISPR interference (CRISPRi) assay to map gene essentiality in phages λ and P1. Published December 4, 2023 in PLoS Biology.
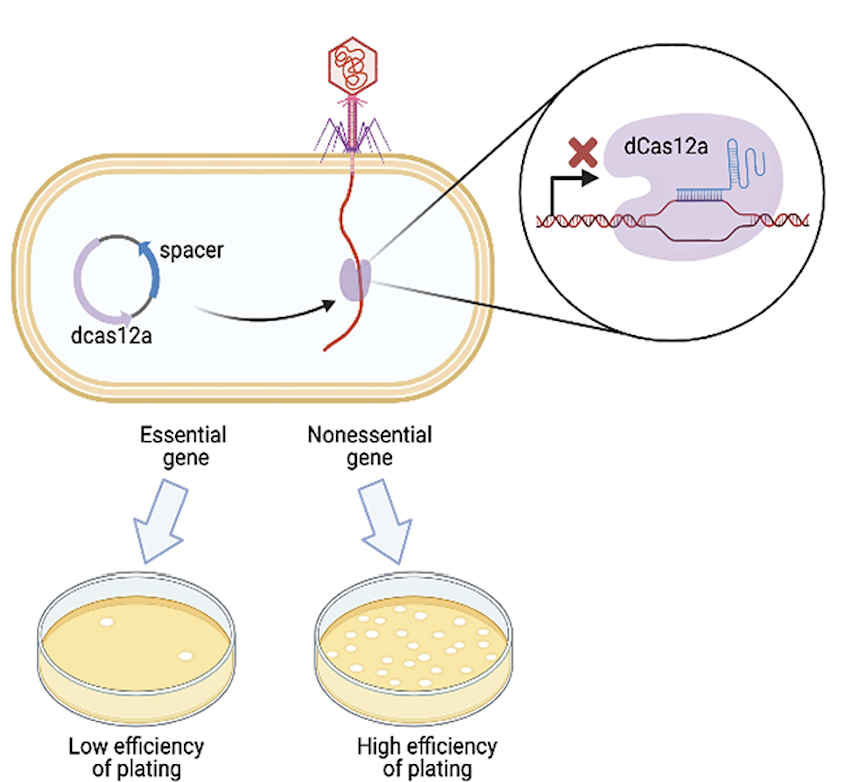
From Piya et al. (2024)
Microbiome transmission mode shapes degree of specialization
Dr. Kyle Meyer and members of the Koskella Lab found that conspecific passaging of leaf microbiomes leads to microbiome specialization, whereas heterospecific passaging leads to microbiome generalism. Published November 28, 2023 in Cell Host & Microbe.
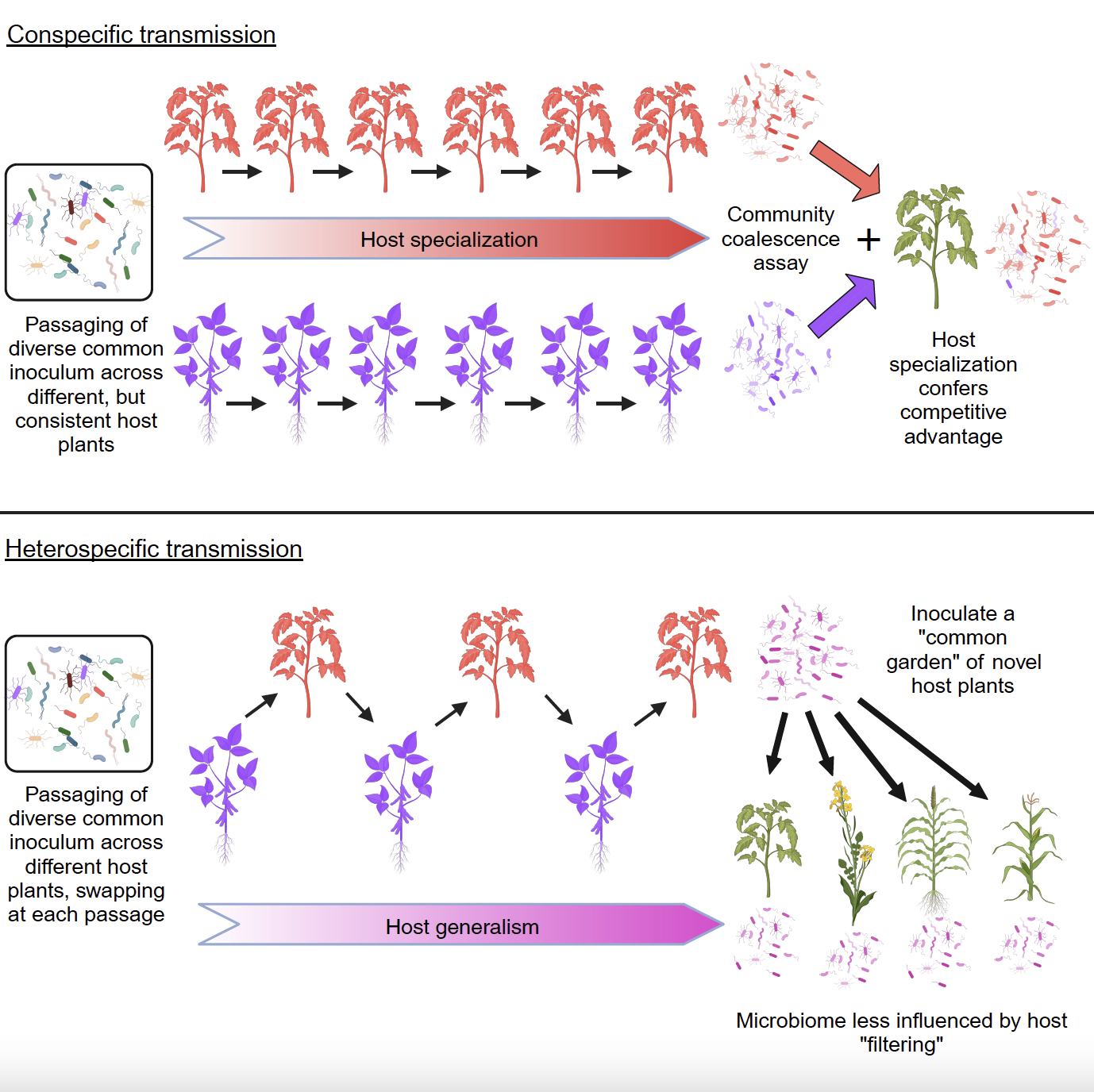
From Meyer et al. (2024)
Upcoming Event: Microbiomes in a Changing Planet
We are excited to announce our upcoming event on Microbiomes in a Changing Planet. This will be a full-day workshop (~9am - 6pm) on November 3 at the International House. Presentations will feature faculty and early career researchers from UCB, LBNL, and other Northern California UC campuses; including a keynote from Dr. Asmeret Asefaw Berhe, Director of the DOE’s Office of Science and Professor of Soil Biogeochemistry at UC Merced.
The day will be divided into three main themes:
- Microbiome response and feedbacks to global change
- Predicting ecosystem and human health impacts
- Translating basic science for microbiome-based solutions
This will be the largest JBIMS event yet! We anticipate spots filling quickly. Reserve your spot here!
We hope to see you there.
Applications are open for the Bay Area RaMP 2023-24 cohort
Interested in a paid post-baccalaureate research training opportunity? Applications are open until May 1, 2023 for the Bay Area RaMP 2023-24 cohort.

Phylosymbiosis in Fish Microbiota
Despite the rapid diversification of morphology and behaviors among pupfish species, gut microbiota related much more closely to the phylogenies of their hosts than to their life history. Reported by the Martin lab (Integrative Biology) in PLOS ONE, September 16, 2022.

Probing Contaminated Environments
From the Terry group (Plant and Microbial Biology): Description of culture-dependent and culture-independent bacterial diversity in contaminated wastewater. Published in Journal of Environmental Management.
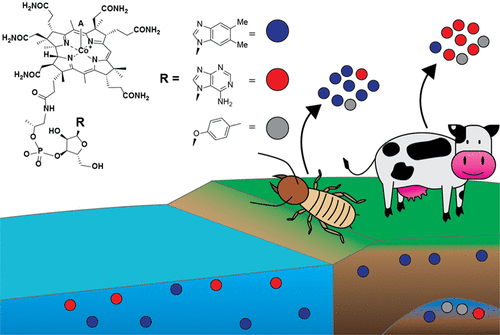
Nutrient Profiles Across Microbiomes
Cobamides are important nutrients in microbial communities that can act as public goods, so cobamide availability in a habitat might shape the structure of microbial interactions. New research from the Taga lab characterizes variation in cobamide composition across insect and mammalian guts and environmental samples. Published August 29, 2022 in Biochemistry.
Bouncing Back After Drought
A new study from the Taylor lab reports that soil fungi are generally more drought-resistant than bacteria, but also that drought disrupts intra- and inter-kingdom correlations in microbial co-occurrence networks. Published July 5, 2022 in Nature Communications.
Microbiome Method Development
Berkeley scientists contributed to two books this summer on characterizing the structure and importance of microbial communities. Learn how to properly run a microbiome GWAS in Methods in Molecular Biology, or read about soil microbiology in desert ecosystems in Microbiology of Hot Deserts. Congratulations to the Coleman-Derr and Northen groups!

Rewriting the Genetic Code
New work from the Banfield lab finds alternative stop codons are widely distributed among phage genomes, including in phages of human and animal gut microbiota. Stop codon has appeared repeatedly in closely related lineages, suggesting it can evolve rapidly. Published May 26, 2022 in Nature Microbiology.
Natural Product Discovery
New paper from the Traxler lab describes the discovery of a new family of antimicrobial compounds, referred to as the dynaplanins. The approach took advantage of natural patterns of antimicrobial production and resistance among soil actinomycetes. Published May 24, 2022 in mBio and selected as a 2022 Editor's Pick.
Analyzing Adaptability
The Chakraborty lab describes how strains within the genus Arthrobacter have adapted to a variety below-ground environments. Published March 30, 2022 in ISME Communications: https://doi.org/10.1038/s43705-022-00113-8